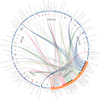
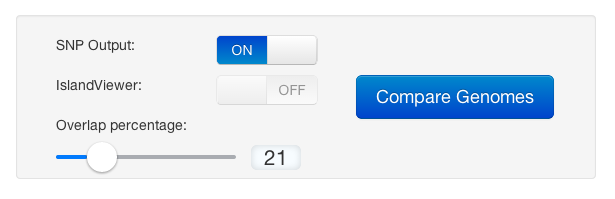
We were trying to keep many options automated so this would be straightforward process, and good for iterations (adding/subtracting genomes, etc). This is what we have boiled the user options down to:
| Option | Description |
|---|---|
| SNP Output | The SNP option simply toggles the track of SNPs to a heatmap. The more SNPs in gene, the "hotter" (more red) it becomes. It calculates the upper limit dynamically. This does use considerable computation resources and increases time to compute. |
| IslandViewer | IV shows predicted genomic islands and virulence islands. This is currently in development and only enabled to certain users for prototype development. Read more about it and development on our Development Blog. |
| Overlap Percentage | The overlap analysis has been included since there will be overlap with varying levels of similarity. A percentage overlap function was added so that the user can specify how much overlap in the genes and backbone is acceptable (i.e., the user sets the desired stringency for calling overlapping data). This step is iterative allowing the user to adjust the analysis to a particular dataset, beginning with the default setting of 0% overlap. |
